Analyzing Urban Heat Islands Using Crowdsourced Air Temperature Data and LCZ4r
Max Anjos
October 25, 2025
Source:vignettes/local_func_modeling_crows.Rmd
local_func_modeling_crows.RmdIntroduction
Urban Heat Islands (UHIs) are a critical environmental issue in cities, where urban areas experience higher temperatures than their rural surroundings. This case study demonstrates how to analyze UHIs using crowdsourced air temperature (Tair) data from Citizen Weather Stations (CWS) and the LCZ4r package in R. We focus on hourly Tair data from May 2023, collected by Netatmo CWS in Berlin, Germany, and available “Crowd-QC” package in R.
Load required packages
# Install and load necessary packages
if (!require("pacman")) install.packages("pacman")
pacman::p_load(remotes, dplyr, sf, tmap, ggplot2, devtools)
library(LCZ4r) # For LCZ and UHI analysis
library(dplyr) # For data manipulation
library(sf) # For vector data manipulation
library(tmap) # For interactive map visualization
library(ggplot2) #For data visualization
library(data.table)
#Install
if (!require("CrowdQCplus")) {remotes::install_github("dafenner/CrowdQCplus")}
library(CrowdQCplus) # For Quality Control of CWS dataDataset
# Retrieve the LCZ map for Berlin using the LCZ Generator Platform
lcz_map <- lcz_get_map_generator(ID = "8576bde60bfe774e335190f2e8fdd125dd9f4299")
# Visualize the LCZ map
lcz_plot_map(lcz_map)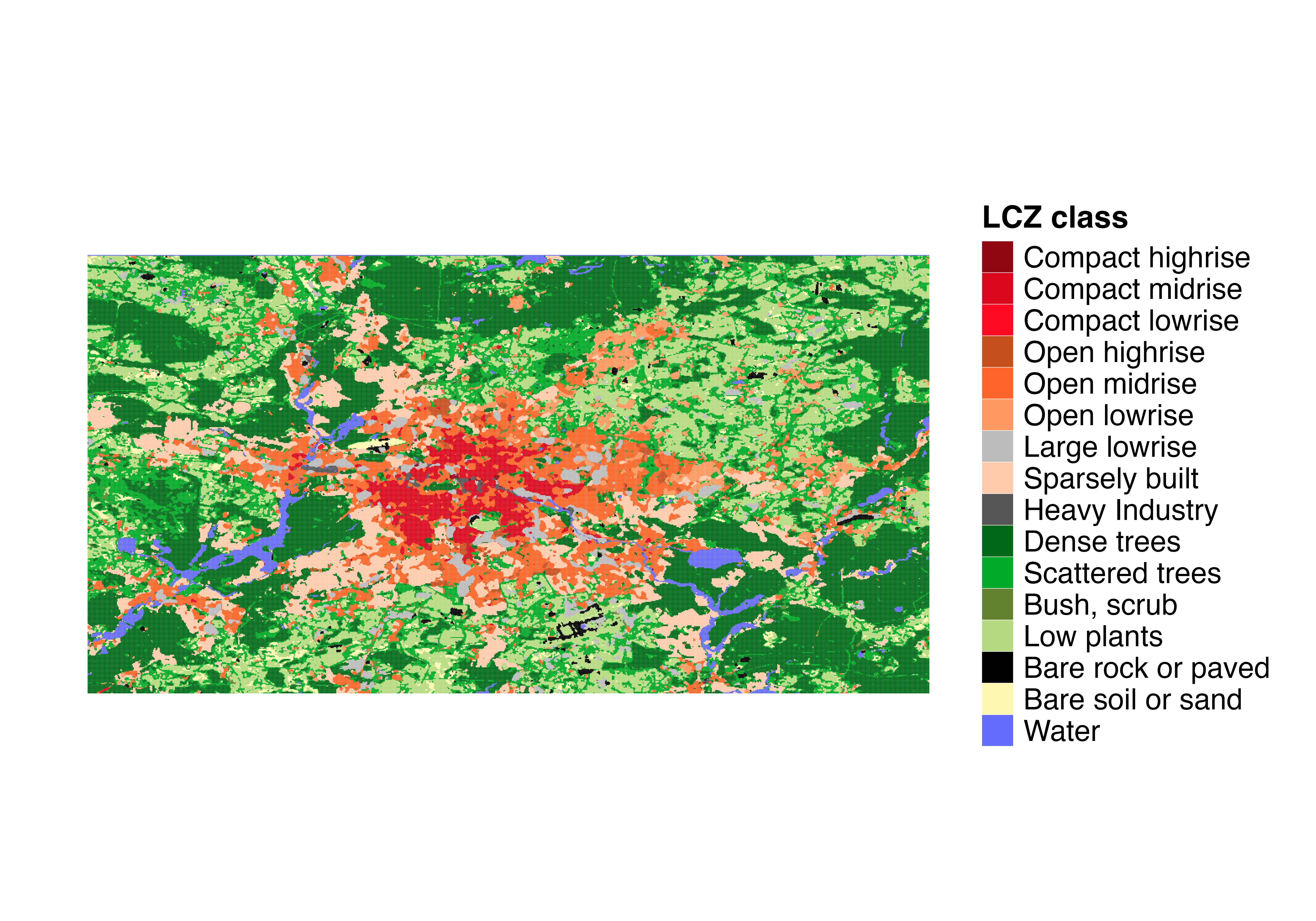
Load and visualize CWS data
# Load sample CWS data for Berlin from the CrowdQCplus package
data(cqcp_cws_data)
head(cqcp_cws_data)
#> p_id time ta lon lat z
#> <int> <POSc> <num> <num> <num> <num>
#> 1: 787964440 2023-05-01 01:00:00 17.42819 13.3926 52.5145 69
#> 2: 787964440 2023-05-01 02:00:00 17.79814 13.3926 52.5145 69
#> 3: 787964440 2023-05-01 03:00:00 16.63420 13.3926 52.5145 69
#> 4: 787964440 2023-05-01 04:00:00 15.56178 13.3926 52.5145 69
#> 5: 787964440 2023-05-01 05:00:00 16.60900 13.3926 52.5145 69
#> 6: 787964440 2023-05-01 06:00:00 17.47491 13.3926 52.5145 69Quality Control of CWS data
To ensure data reliability, we applied the quality control (QC) method developed by Fenner et al. (2021) using the CrowdQCplus package. This process filters out statistically implausible readings, improving data accuracy.
After applying QC, multiple columns are generated. For air temperature interpolation, we selected the “ta_int” column from the “data_qc” dataframe, as it represents temperature values that successfully passed all quality control steps.
# Perform data padding and input checks
data_crows <- cqcp_padding(cqcp_cws_data)
data_check <- cqcp_check_input(data_crows)
#> +++++++++++++++++++++++++++++
#> + CrowdQC+ input data check +
#> +++++++++++++++++++++++++++++
#> Check 1a - Required columns:
#> OK
#> Check 1b - Optional columns:
#> OK
#> Check 1c - Column data types:
#> OK
#> Check 2 - Temporal coverage:
#> OK
#> Check 3 - Regularity:
#> OK
#> Check 4 - Geographical extent:
#> OK
#> Check 5 - Number of stations:
#> OK
#>
# Apply quality control
if (data_check) {
data_qc <- cqcp_qcCWS(data_crows) # QC
}
head(data_qc)
#> Key: <p_id, time>
#> time p_id lon lat z ta m1
#> <POSc> <int> <num> <num> <num> <num> <lgcl>
#> 1: 2023-05-01 01:00:00 1312380 12.95865 52.61623 28.85817 14.97525 TRUE
#> 2: 2023-05-01 02:00:00 1312380 12.95865 52.61623 28.85817 NA FALSE
#> 3: 2023-05-01 03:00:00 1312380 12.95865 52.61623 28.85817 NA FALSE
#> 4: 2023-05-01 04:00:00 1312380 12.95865 52.61623 28.85817 14.71264 TRUE
#> 5: 2023-05-01 05:00:00 1312380 12.95865 52.61623 28.85817 15.82922 TRUE
#> 6: 2023-05-01 06:00:00 1312380 12.95865 52.61623 28.85817 NA FALSE
#> m2 m3 m4 m5 isolated ta_int o1 o2 o3
#> <lgcl> <lgcl> <lgcl> <lgcl> <lgcl> <num> <lgcl> <lgcl> <lgcl>
#> 1: TRUE TRUE TRUE FALSE FALSE NA FALSE FALSE FALSE
#> 2: FALSE FALSE FALSE FALSE FALSE NA FALSE FALSE FALSE
#> 3: FALSE FALSE FALSE FALSE FALSE NA FALSE FALSE FALSE
#> 4: TRUE TRUE TRUE FALSE FALSE NA FALSE FALSE FALSE
#> 5: TRUE TRUE TRUE FALSE FALSE NA FALSE FALSE FALSE
#> 6: FALSE FALSE FALSE FALSE FALSE NA FALSE FALSE FALSE
# Display QC output statistics
n_data_qc <- cqcp_output_statistics(data_qc)
#> ++++++++++++++++++++++++++++++
#> + CrowdQC+ output statistics +
#> ++++++++++++++++++++++++++++++
#> Raw data: 298706 values, 624 stations
#> QC level m1: 275666 values (= 92.29 % of raw data), 594 stations
#> QC level m2: 260503 values (= 87.21 % of raw data), 590 stations
#> QC level m3: 258451 values (= 86.52 % of raw data), 577 stations
#> QC level m4: 230233 values (= 77.08 % of raw data), 528 stations
#> QC level m5: 34076 values (= 11.41 % of raw data), 242 stations
#> QC level o1: 40319 values (= 13.50 % of raw data), 242 stations
#> QC level o2: 8216 values (= 2.75 % of raw data), 30 stations
#> QC level o3: 3433 values (= 1.15 % of raw data), 5 stationsDive into CWS LCZs
Diurnal Cycle of Air Temperature
We examined the diurnal cycle of air temperature for a specific day (May 1, 2023) across different LCZs.
cws_ts <- lcz_ts(lcz_map, data_frame = data_qc,
var = "ta_int", station_id = "p_id",
time.freq = "hour",
extract.method = "two.step",
year = 2023, month = 5, day = 1,
by = "daylight")
cws_ts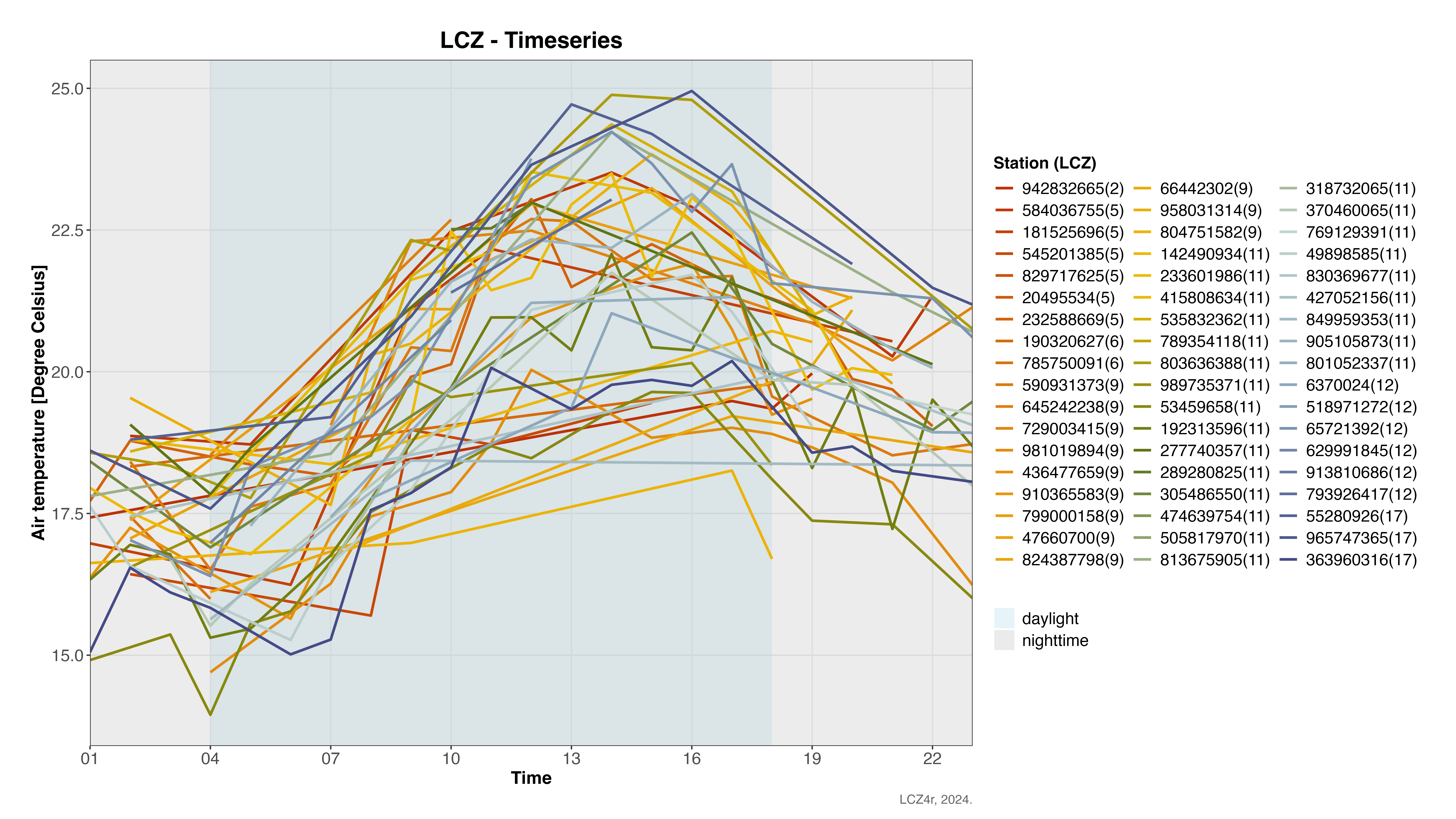
Urban heat island Analysis
Intensity
We calculated hourly UHI intensities for May 2023 using the LCZ method, which automatically selects urban and rural temperatures based on LCZ classes. The highest UHI intensity observed was 3.7°C on May 13 at 16:00.
cws_uhi <- lcz_uhi_intensity(lcz_map, data_frame = data_qc,
var = "ta_int", station_id = "p_id",
time.freq = "hour",
extract.method = "two.step",
year = 2023, month = 5,
method = "LCZ")
cws_uhi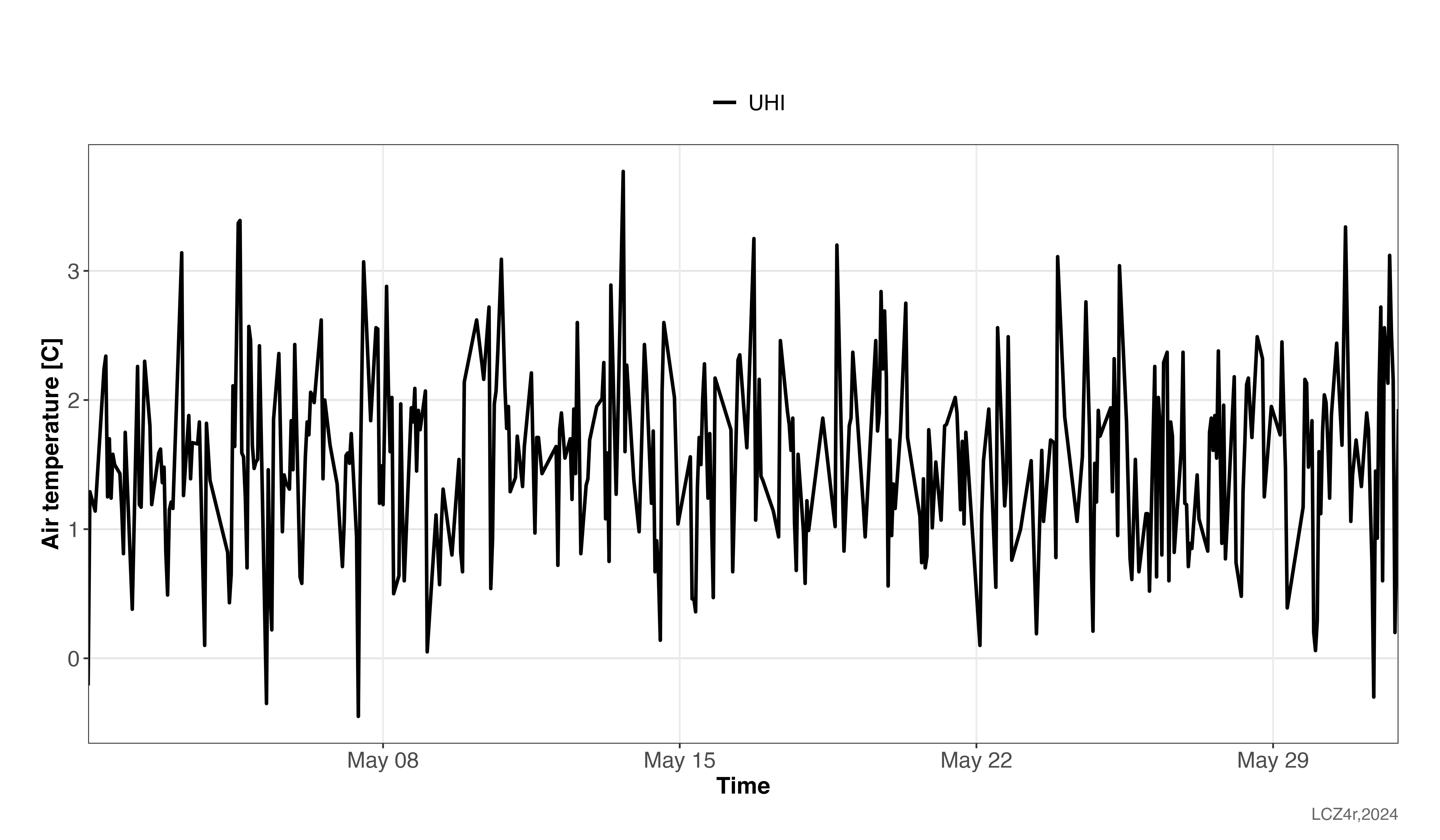
UHI mapping
We modeled the UHI for May 13, 2023, at 16:00, using LCZ-based spatial interpolation.
# Map air temperatures for 2023-05-13 at 16h.
my_interp_map <- lcz_interp_map(
lcz_map,
data_frame = data_qc,
var = "ta_int",
station_id = "p_id",
sp.res = 100,
tp.res = "hour",
year = 2023, month = 5, day = 13, hour = 16
)
# Customize the plot with titles and labels
lcz_plot_interp(
my_interp_map,
title = "Thermal field - CWS data",
subtitle = "Berlin - 2023-05-13 at 16h",
caption = "Source: LCZ4r, 2024.",
fill = "[ºC]")
# Map air temperatures for 2023-05-13 at 16h.
my_anomaly_map <- lcz_anomaly_map(
lcz_map,
data_frame = data_qc,
var = "ta_int",
station_id = "p_id",
sp.res = 100,
tp.res = "hour",
year = 2023, month = 5, day = 13, hour = 16
)
# Customize the plot with titles and labels
lcz_plot_interp(
title = "Thermal anomaly field - CWS data",
subtitle = "Berlin - 2023-05-13 at 16h",
caption = "Source: LCZ4r, 2024.",
fill = "[ºC]",
palette = "bl_yl_rd")
Evaluate UHI map
We evaluated the interpolation accuracy using Leave-One-Out Cross-Validation (LOOCV) and calculated metrics such as RMSE, MAE, and sMAPE.
# Evaluate the interpolation
df_eval <- lcz_interp_eval(
lcz_map,
data_frame = data_qc,
var = "ta_int",
station_id = "p_id",
year = 2023, month = 5, day = 13,
LOOCV = TRUE,
extract.method = "two.step",
sp.res = 500,
tp.res = "hour",
vg.model = "Sph",
LCZinterp = TRUE
)Evaluation metrics
Based on the table results, we calculate evaluation metrics (RMSE, MAE MAPE) to quantify uncertainties.
Correlation and Residual Analysis
We analyzed the correlation between observed and predicted values and examined residuals by LCZ.
# Correlation plot with regression equation and R2
p1 <- ggplot(df_eval, aes(x = observed, y = predicted)) +
geom_point(alpha = 0.5, color = "blue") + # Scatter points
geom_smooth(method = "lm", color = "red", se = FALSE) + # Regression line
stat_density2d(aes(fill = ..level..), geom = "polygon", alpha = 0.3) + # Density contours
scale_fill_viridis_c() + # Viridis color scale for density
stat_poly_eq(
aes(label = paste(..eq.label.., ..rr.label.., sep = "~~~")),
formula = y ~ x,
parse = TRUE,
label.x.npc = "left", # Position of the equation
label.y.npc = "top"
) + # Add regression equation and R2
labs(
title = "",
x = "Observed values",
y = "Predicted values"
) +
theme_minimal(base_size = 14) # Minimal theme for elegance
# Add marginal histograms
p1_with_marginals <- ggExtra::ggMarginal(p1, type = "histogram", fill = "gray", bins = 30)
# Print the plot
p1_with_marginals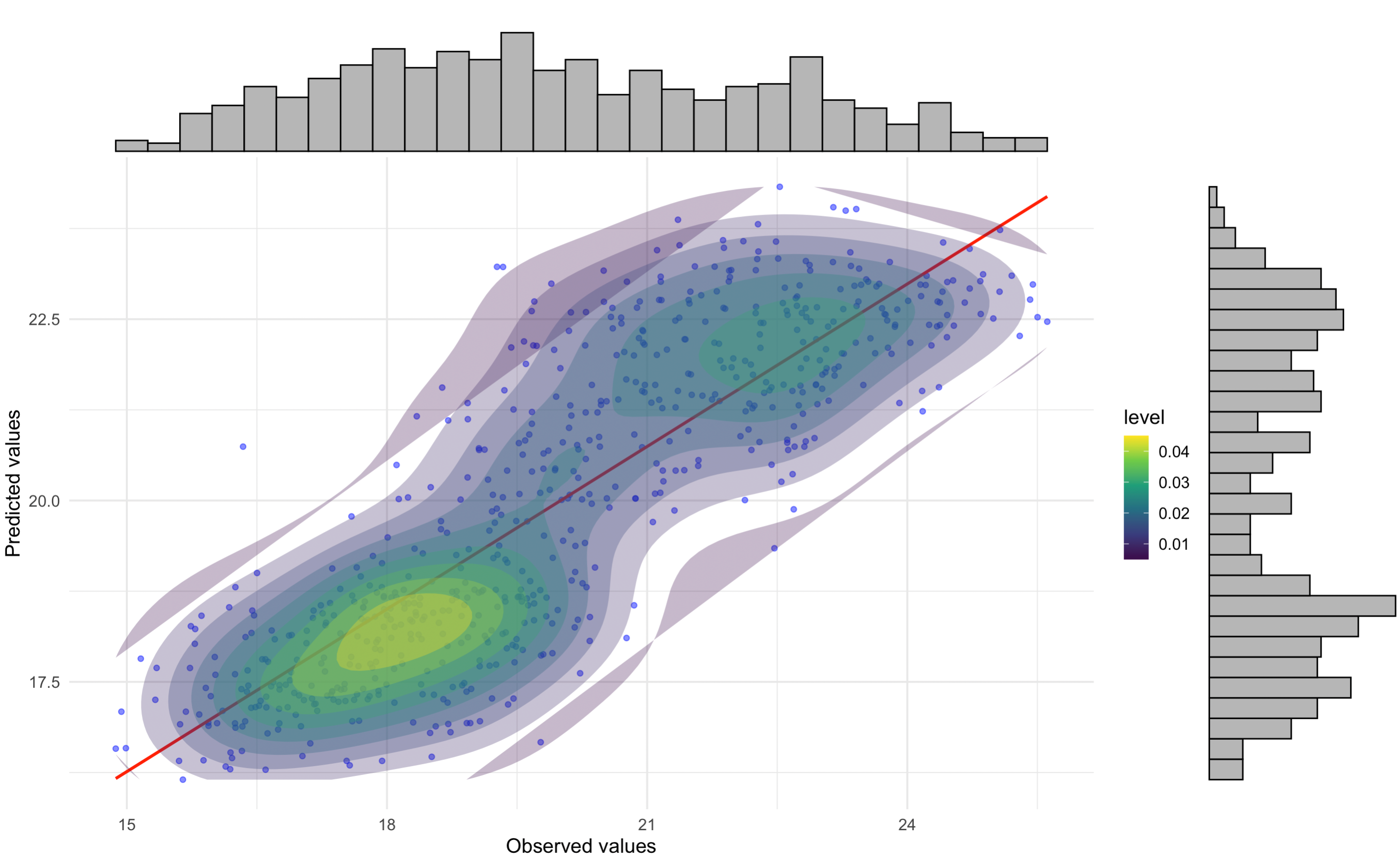
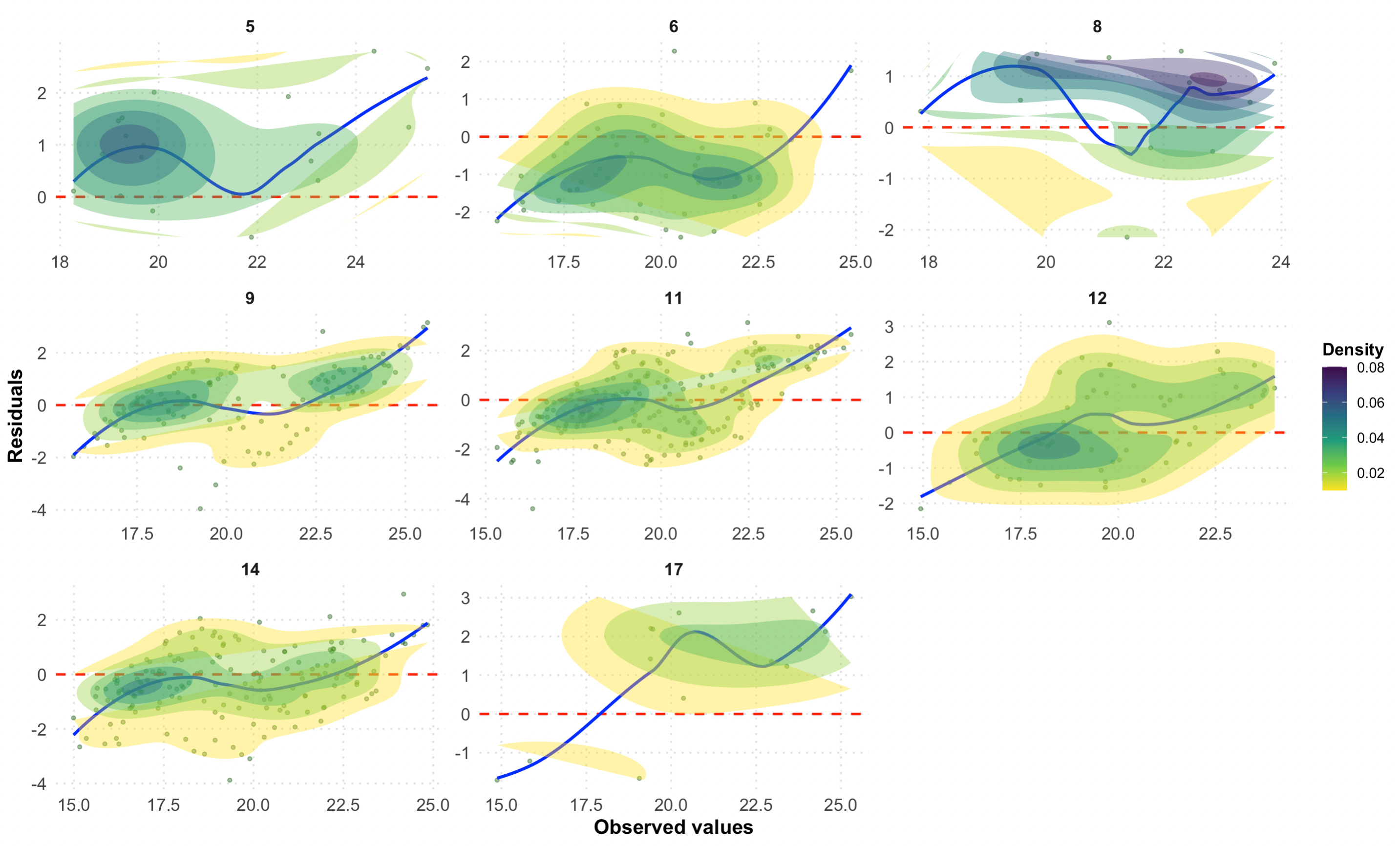
# Residuals plot by LCZ
p2 <- ggplot(df_eval, aes(x = observed, y = residual)) +
# Scatter points with transparency for better visibility
geom_point(alpha = 0.4, color = "darkgreen", size = 1) +
# Horizontal reference line at 0
geom_hline(yintercept = 0, linetype = "dashed", color = "red", size = 0.8) +
# Loess smoothing line for trend
geom_smooth(method = "loess", color = "blue", se = FALSE, size = 1) +
#Density contours for point concentration
stat_density2d(aes(fill = ..level..), geom = "polygon", alpha = 0.4) +
scale_fill_viridis_c(option = "D", direction = -1) + # Viridis color scale
# Facet by LCZ with free scales
facet_wrap(~ lcz, scales = "free", ncol = 3) +
# Labels and titles
labs(
title = "",
x = "Observed values",
y = "Residuals",
fill = "Density"
) +
# Customize
theme_minimal(base_size = 14) +
theme(
plot.title = element_text(size = 16, face = "bold", hjust = 0.5), # Centered bold title
axis.title = element_text(size = 14, face = "bold"), # Bold axis titles
axis.text = element_text(size = 12), # Axis text size
strip.text = element_text(size = 12, face = "bold"), # Facet labels
legend.position = "right", # Place legend on the right
legend.title = element_text(size = 12, face = "bold"), # Bold legend title
legend.text = element_text(size = 10), # Legend text size
panel.grid.major = element_line(color = "gray90", linetype = "dotted"), # Subtle gridlines
panel.grid.minor = element_blank()
)
# Print the plot
p2